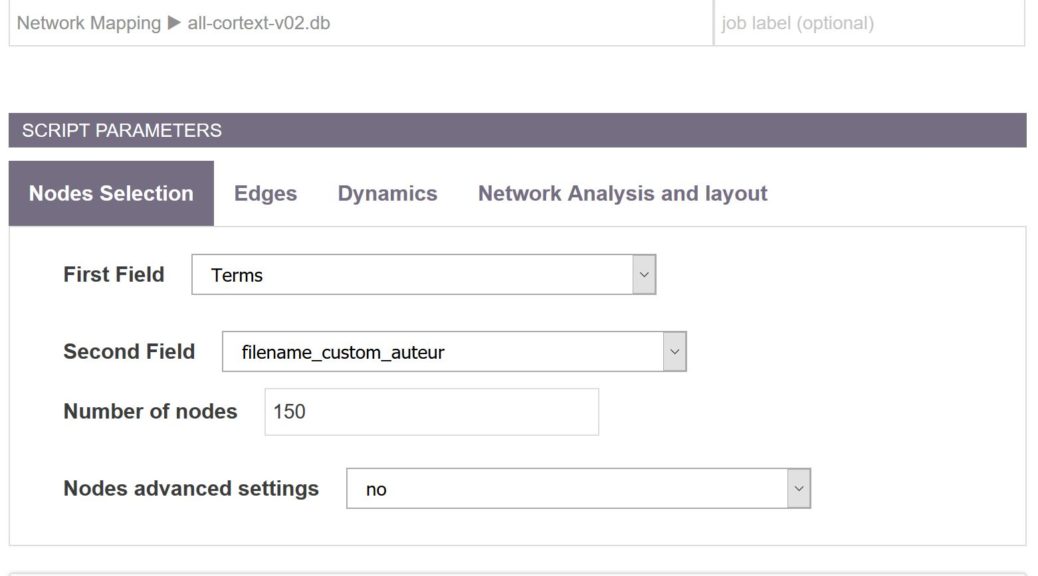
This page describes the main options available under the node panel of the mapping script.
Heterogeneous and homogeneous networks
The underlying rationale behind this analysis is to use a very systematic and symmetric perspective that allow users to produce heterogeneous networks featuring any couple of nodes types. One can mix any two fields: for example from an ISI extraction: authors and their keywords, journals of publication and the journals they cite (journal level inter-citation network), extracted terms and cited journals, countries and cited references, years and terms, etc.
Of course it is also possible (and even advised in the first place!) to produce traditional homogeneous networks like: co-authorship, co-words, or co-citation networks.
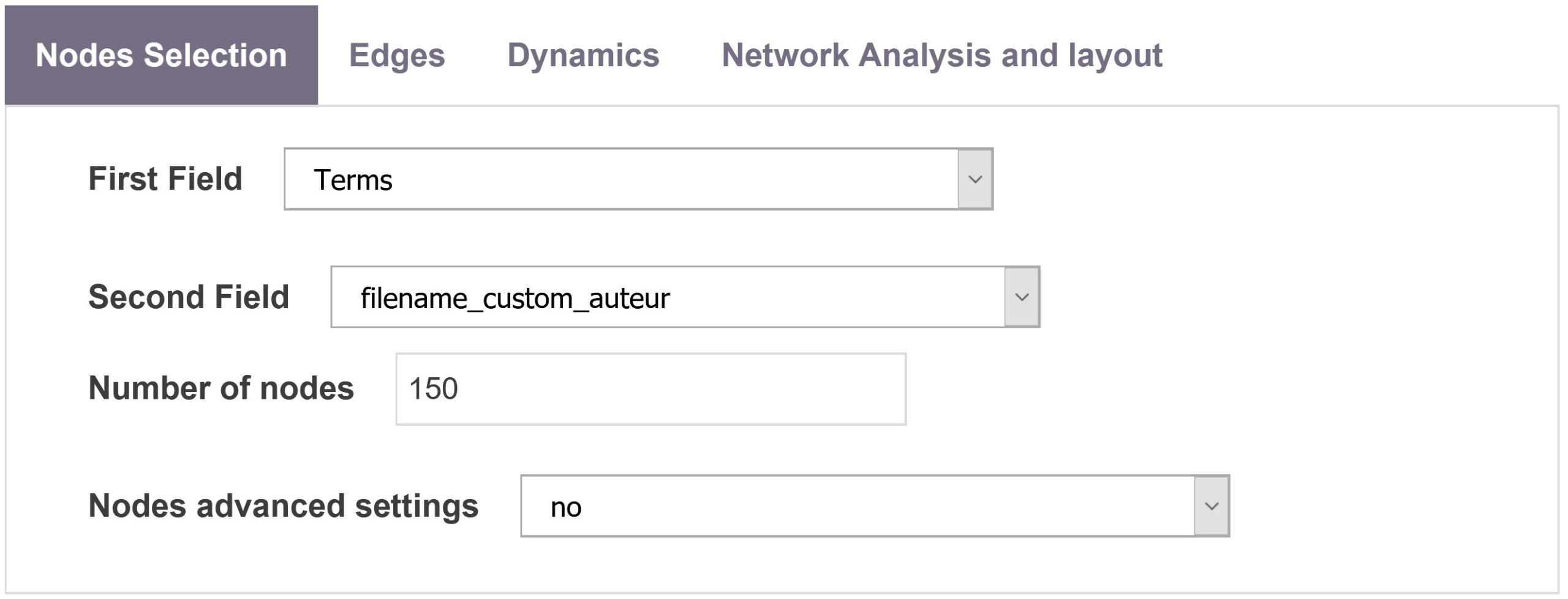
Choose first field and second field
To select the fields you wish to map, simply select the fields (first field and second field) in the node selection tab.
Number of nodes
The number of nodes to be mapped can also be easily tuned. For computational reasons, the maximal number of nodes is still limited to 500. The nodes are selected according to their frequency at each time period. For example when mapping a co-authorship network, choosing 50 top items will produce the collaboration network between the 50 most productive authors (in terms of articles production) at each time period. For a research lab vs keywords map, 50 most productive research labs will be mapped along with the 50 most frequent keywords.
Nodes advanced settings
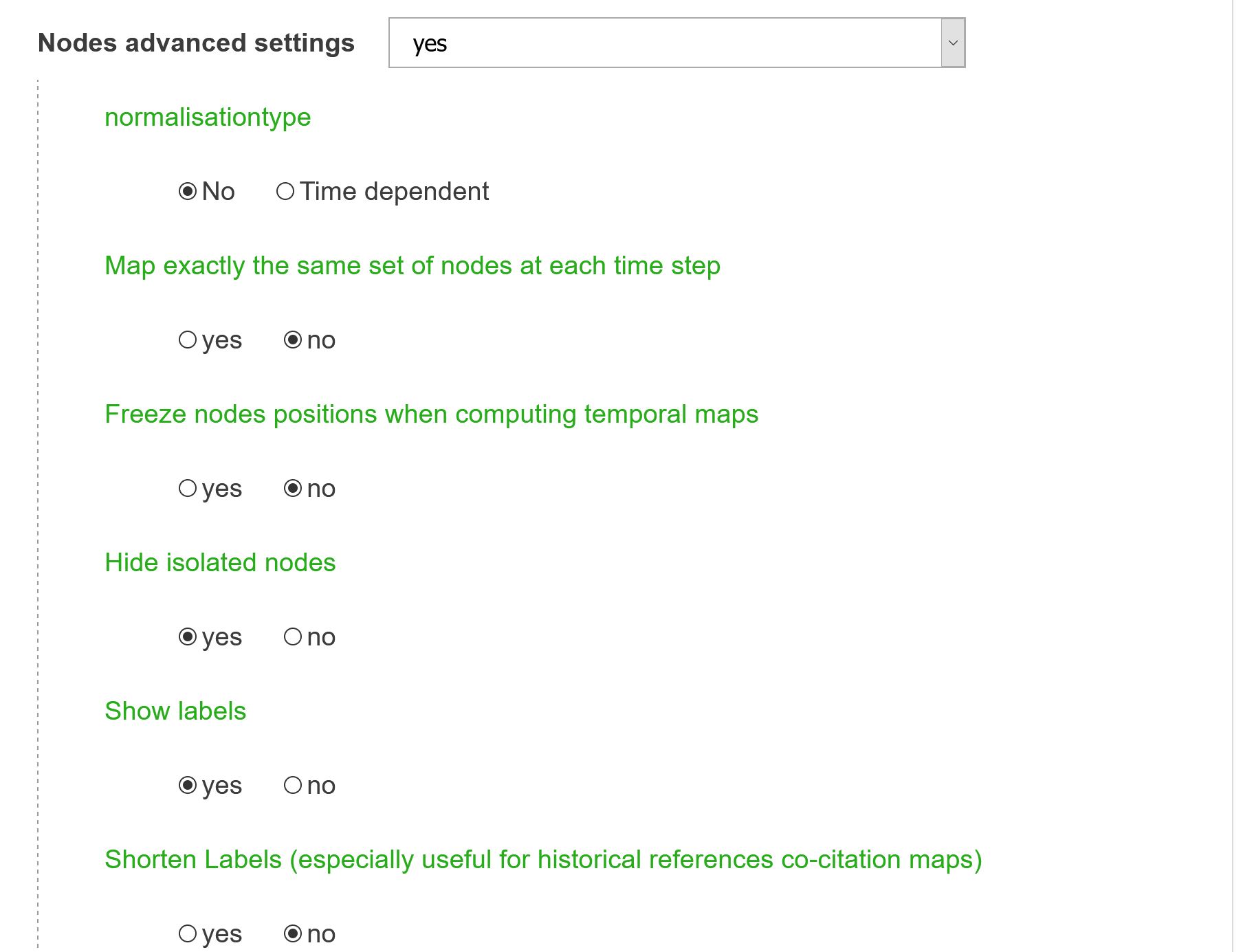
normalisationtype
The top N items are not based on their overall frequency (which is the default behavior). If time dependent is checked, for each time step (typically year) the proportion of articles mentioning every item is computed. Those proportions are added over all the possible time steps resulting in a score which allows to rank all the entities and select the top N nodes which will be mapped.
Map exactly the same set of nodes at each time step
Fixes the set of nodes (only pertinent when several time periods are set). By default, N most frequent entities will be selected at each time period, likely resulting in only partially overlapping nodes being mapped. If you check this option, the N most frequent nodes over the whole time period will be selected.
Freeze nodes positions when computing temporal maps
When computing successive maps, nodes positions are recomputed at each time step by default. Select this option if you prefer to freeze their position starting from the ones computed during the first time period.
Hide isolated nodes
By default, nodes with degree zero are not shown, you can deactivate this option to reveal them (they should then appear on the left side of the map)
Show labels
Names of nodes will be shown or not
Shorten labels
Names of nodes are reduced to set size.
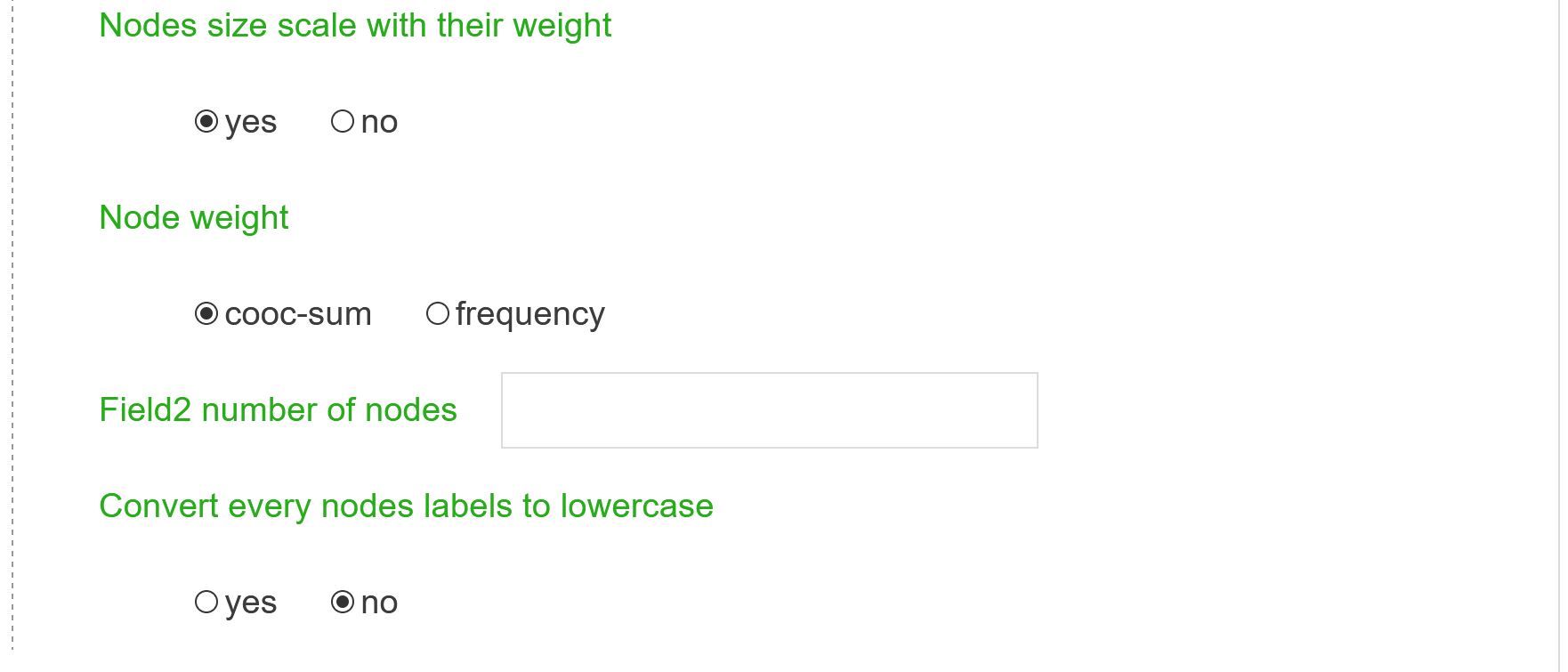
Nodes size scale with their weight
Node weight values are used to determine the size of nodes. If no: all nodes will have the same size.
Node weight
Node weight defined by co-occurrence sum or frequency
Field2 number of nodes
If you wish a different number of nodes for the second field. The first field will remain the same.
Convert every nodes labels to lowercase
Convert nodes labels to lowercase. Useful for Cited and Citing Journals networks to build inter-citation network and merged Cited and Citing Journals labels.