Once you have collected data (see Data formats section for more information about data supported by CorText Manager), you should first zip the file(s) that compose your corpus into a single zip archive before you can upload it into CorText Manager.
The zip archive should be performed, regardless of the format of your original data (e.g. a set of txt files downloaded from Web Of Science, pdf files, a database…) and of the number of files that are part of your corpus (e.g. if your corpus is composed of only one csv file, this one must also be zipped).
The mandatory step to be able to mobilize analysis scripts in CorText manager is then to run the “Data Parsing” script (see this page for an explanation of what the ”Data Parsing” script does). “Data Parsing” script can only process one unique zip file.
Upload process
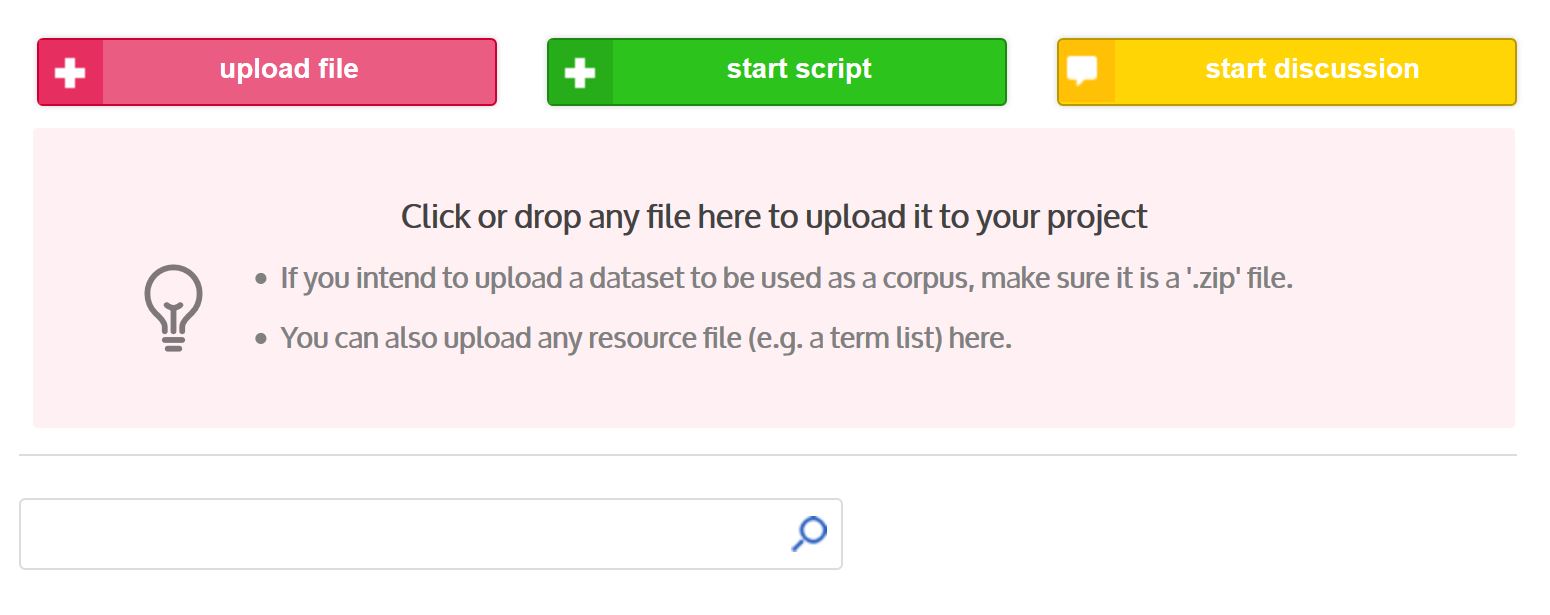
Click on the “+ upload file” menu. You can then drag and drop the zip file or enter the path of the zip containing your corpus.
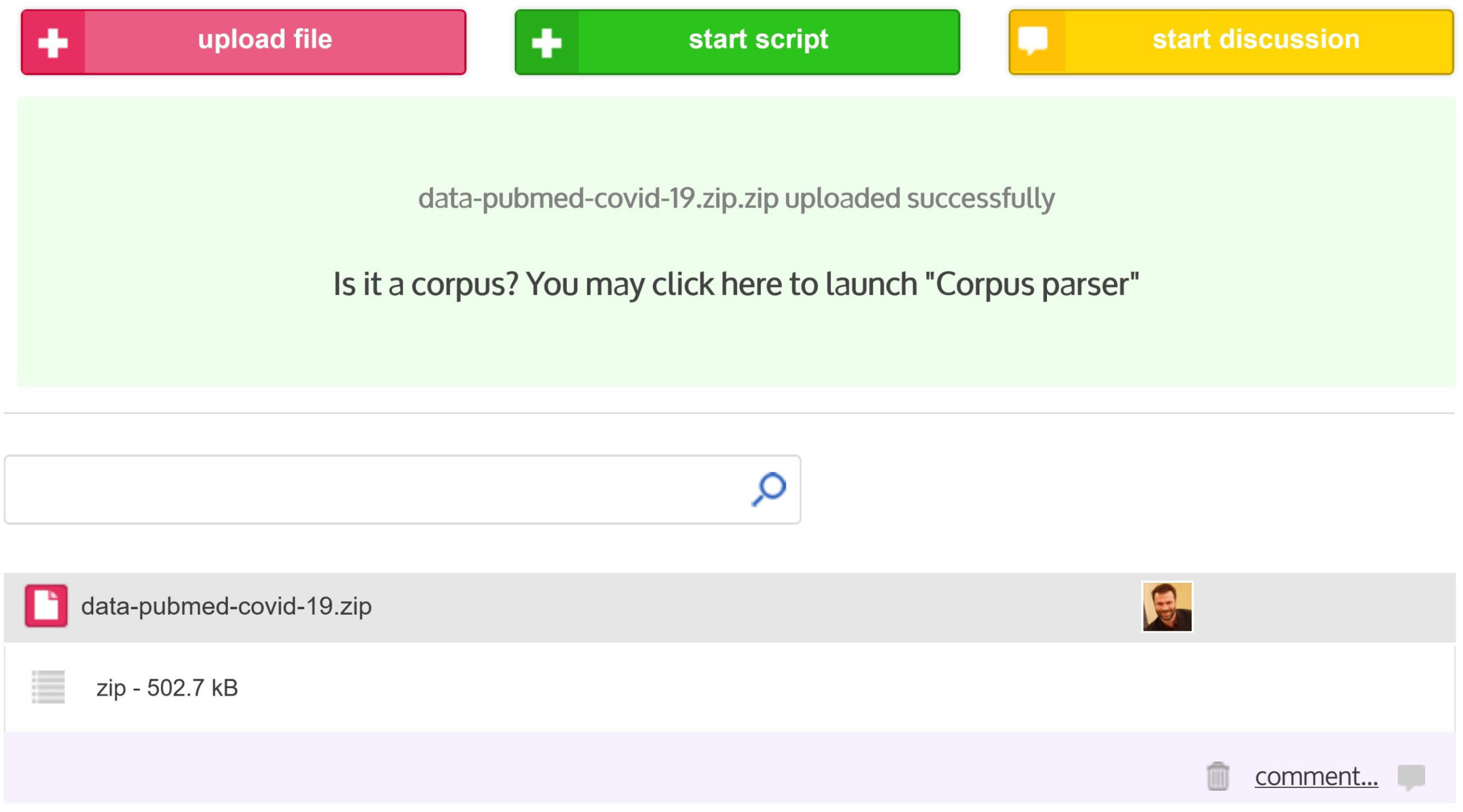
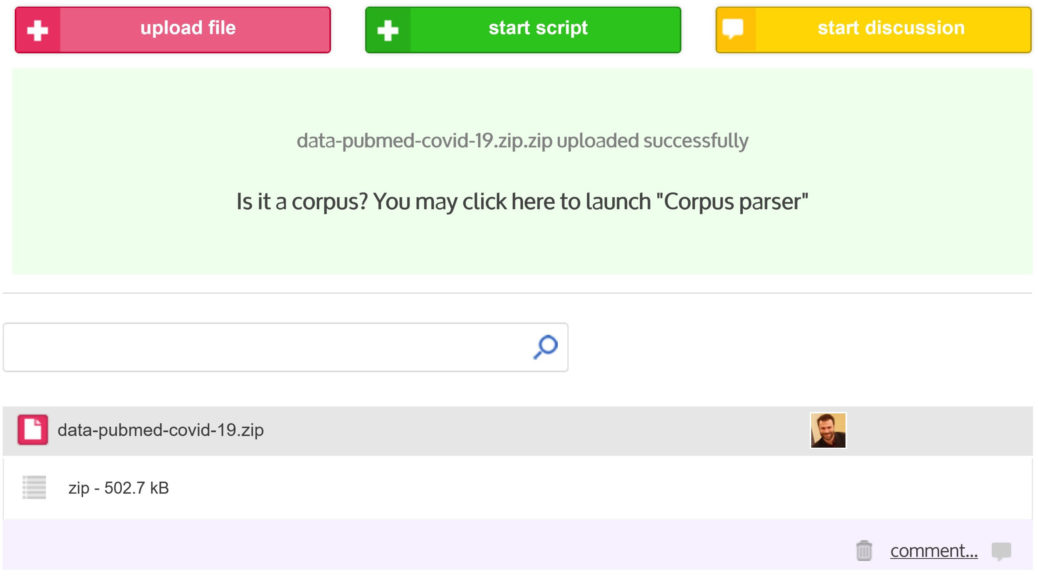
 At the end of the upload progress bar (located in the upper left corner of the project dashboard), if the upload is a success (“File uploaded” will appear), click in the green box and you will be automatically redirected on the “Data Parsing” script window.
At the end of the upload progress bar (located in the upper left corner of the project dashboard), if the upload is a success (“File uploaded” will appear), click in the green box and you will be automatically redirected on the “Data Parsing” script window.
At anytime you can also access to the “Data Parsing” script using the common way: “+ start script” then selecting “Data Parsing”.
An entry will also be created in the dashboard containing the zip file.