I understand that PubMed no longer uses the xml format and one therefore has to download PubMed references in .nbib format, import them to Zotero (which turns out to be quite buggy), re-export them in RIS format and upload them to Cortext. Two questions:
(1) Lionel mentioned that he was working on a new PubMed parser: is this still the case and, if so, when should the new parser become available?
(2) When I upload and parse a PubMed database and I select Demography, main parameters, I’m asked to select which variables, and I see a list of acronyms, some of which are quite mysterious: would it be possible to have a full-name list of the variables to which these acrinyms correspond?
Dear Albertoc,
- Please have a look to this related page on the CorText Q&A forum : https://docs.cortext.net/question/pubmed-and-xml-extraction/
We are not specialists of nbib format, neither of the conversion between nbib to a ris using Zotero, but for the nbib fields definition, you can read https://www.nlm.nih.gov/bsd/mms/medlineelements.html I do not know how Zotero translate the fields names during the conversion. - True, we are working on a new nbib parser, which will be released during the next months, first semester of 2022. Stay tuned.
I hope it helps
L
Dear Lionel,
Many thanks for the quick reply. I’m will stay tuned for the new nbib parser!
Happy Holidays,
alberto
Your welcome Alberto! We will let you know, here.
Happy winter Holiday.
L
Hello,
Is there any update concerning a parser for a PubMed database?
Btw, I noticed that the U of Meryland has developed a script to use PubMed PMIDs to download PubMed references in xml format.
https://answers.hshsl.umaryland.edu/faq/318581
Thanks,
alberto
Dear Alberto,
Many thanks for the follow-up!
Indeed, we have developed and deployed a parser for the nbib format, used in particular by PubMed. It has been a few months, so it is not yet documented.
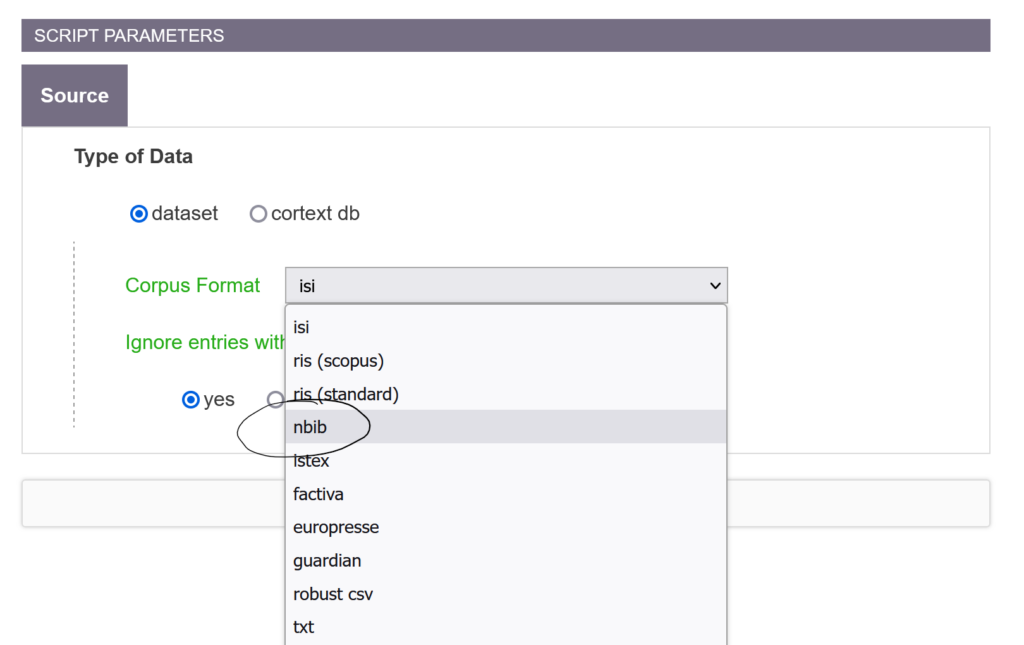
Don’t hesitate to use it and to give us feedback! It will be welcome.
Kind regards,
L
